A new genetic marker type recently termed SNPSTR (pronounced ‘snipster’) is being established for the species under investigation. The name derives from its structure: A microsatellite (STR, short tandem repeats) forms the center, and the flanking regions contain at least one sequence variant (SNP, single nucleotide polymorphism).
The SNPSTR technology combines short DNA motifs that are repeated several times with patterns of punctual individual deviations (SNPs) in the genetic material. Parentage analysis is enabled by the information gained from the microsatellites, while the information from the SNPs allows the determination of the population origin.
When analyzing animal populations or breeding, SNPSTRs offer significant advantages over examining microsatellites or SNPs separately. Due to the difference in mutation rates between SNPs and STRs, the information content per gene locus significantly increases, requiring fewer markers for a reliable match or mismatch. Additionally, the flanking regions of microsatellites are highly conserved, and with the SNPs and STRs located in close proximity (approximately 500 bp), recombination is unlikely. This allows the development of a haplotype consisting of two markers, which also provides higher discriminative power.
Our objective is to develop a set of approximately 20 SNPSTR markers per selected species, which will allow the reliable determination of the population origin and paternity of the animal in question.
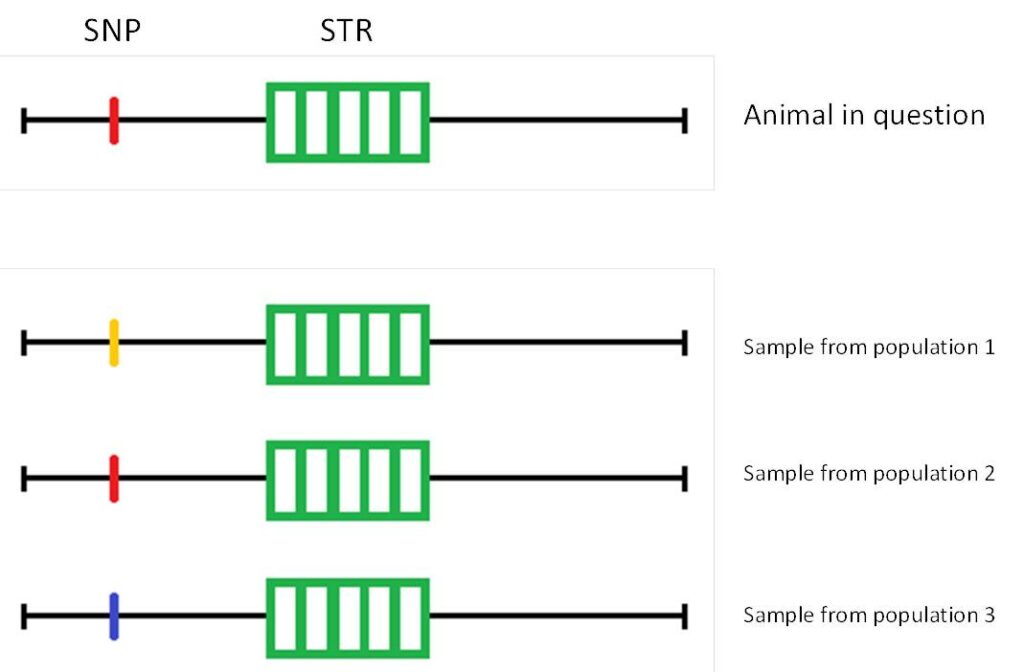
Figure 1: A SNPSTR marker with a microsatellite (5 repeats of the DNA motif) and a SNP. Based on the SNP, the individual in question can be explicitly assigned to population 2. In the case of a parentage analysis, the STRs would be used.(Sketch based on Knieps, 2020, Bachelor thesis)

Laboratory staff examine DNA material at the Alexander Koenig Zoological Research Museum.
